|
组长:冯婕 研究员、博士生导师 |
|
研究组研究方向
病原细菌多重耐药机制及噬菌体防治策略研究
研究组研究内容及意义
本研究组致力于病原菌耐药机制以及噬菌体防治策略的研究。针对临床和环境耐药菌,揭示其耐药机制,发现新的耐药基因或机制,阐明耐药基因的转移和传播机理,为控制细菌耐药性提供理论基础。同时,在深入了解病原菌耐药的背景下,以能够消除多重耐药菌的噬菌体为研究对象,建立裂解不同血清型和序列型病原菌的噬菌体资源库,揭示病原菌受体与噬菌体受体结合蛋白的相互关系,进一步研究病原菌防御和噬菌体反防御的新机制,为解决噬菌体窄谱和抗性问题提供新思路、新策略。最终,病原菌耐药机制和噬菌体防治策略的研究将为控制日益严重的细菌耐药问题奠定理论基础和提供技术支撑。 |
|
研究组长
研究组长:冯婕 电子邮件:fengj#im.ac.cn(请将#替换成@) 通讯地址:北京市朝阳区北辰西路1号院3号,中国科学院微生物研究所 邮政编码:100101 |
|
主要学习及工作经历
2002-2006,中国科学院微生物研究所,博士 2006-2009,加拿大Laval University, Infectious Disease Research Centre,博士后 2009-2016,中国科学院微生物研究所,微生物资源前期开发国家重点实验室,青年研究组长,副研究员 2017-至今,中国科学院微生物研究所,微生物资源前期开发国家重点实验室,研究组长,研究员,博士生导师 |
|
学术兼职情况
现任中国分析微生物学会专业委员会委员、中国噬菌体技术专业委员会委员、微生物学通报编委。
|
|
研究团队
2023年实验室春游
2016年实验室滑雪活动
2022年实验室毕业合影
工作人员
王超,博士,副研究员 研究方向:细菌耐药及噬菌体防治研究 wangchao@im.ac.cn
张刚,博士,副研究员 研究方向:病原细菌耐药机制以及基于噬菌体改造的耐药性防治研究 zhanggang@im.ac.cn
宋宇琴,博士,助理研究员 研究方向:基于生物信息学的细菌耐药和噬菌体抗性研究 songyq@im.ac.cn
魏大伟,博士后 研究方向:病原菌耐药和毒力机制研究 imcas.daweiwei@gmail.com
在读研究生 博士生: 姚仕钢,2020年入学 穆永其,2021年入学 荆世松,丁子煊(联培)2022年入学 黄江庆,2023年入学
硕士生: 李毅,杨丽丽(联培),2021年入学 王新宇,田雪茹(联培),2022年入学 孙文悦,宋怡瑶(联培),2023年入学 |
|
代表性论文(#共同一作,*通讯作者)
1. Yao Shigang#, Wu Xinyi#, Li Y, Song Y, Wang C, Zhang Gang*, Feng Jie*. Harnessing the native type I-F CRISPR-Cas system ofAcinetobacter baumanniifor genome editing and gene repression. Int J Antimicrob Agents. 2023 Sep 4;62(5):106962. 2. Zeng Yuan#, Song Yuqin#, Cui Lanqing#, Wu Q, Wang C, Coelho AC, Zhang G, Wei D, Li C, Zhang J, Corbeil J, Li Yun*, Feng Jie*. Phylogenomic insights into evolutionary trajectories of multidrug resistantS. pneumoniaeCC271 over a period of 14 years in China. Genome Med. 2023 Jul 4;15(1):46. 3. Zhang Gang#, Yao Shigang#, Liu Yunan, Fang Hailing, Song Yuqin, Wang,Chao Wei Dawei, Feng Jie*. Systematic discovery of a new catalogue of tyrosine-type integrases in bacterial genomic islands. Appl Environ Microbiol. 2023 Feb 28;89(2):e0173822. 4. Yao Shigang, Wei Dawei, Tang Na, Song Yuqin, Wang Chao, Feng Jie*, Zhang Gang*. Efficient suppression of natural plasmid-borne gene expression in carbapenem-resistantKlebsiella pneumoniaeusing a compact CRISPR interference system. Antimicrob Agents Chemother. 2022 Nov 15;66(11):e0089022. 5. Yang Jing#, Li Yi#, Tang Na#, Li J#, Zhou J, Lu S, Zhang G, Song Y, Wang C, Zhong J, Xu J*, Feng Jie*. The human gut serves as a reservoir of hypervirulent Klebsiella pneumoniae. Gut Microbes. 2022 Jan-Dec;14(1):2114739. 6. Wei Dawei#, Wong N#, Song Y, Zhang G, Wang C, Li J, Feng Jie*. IS26veers genomic plasticity and genetic rearrangement toward carbapenem hyperresistance under sublethal antibiotics. mBio. 2022 Feb 8;13(1):e0334021. 7. Tang Na#, Hu J#, Zhao Y, Song Y, Wang C, Zhang G, Wei D, Fang H, Li C, Jia R, Feng Jie*. In vivo evolution of carbapenem resistance in hypervirulentKlebsiella pneumoniaein a patient undergoing long-term treatment. J Antimicrob Chemother. 2022 Feb 2;77(2):531-533. 8. Zhang Gang#,Li Jianjuan#, Ai Guomin, He Jialiang, Wang Chao*, Feng Jie*. A new intrinsic aminoglycoside 6'-N-acetyltransferase subclass, AAC(6')-III, inBurkholderiapseudomallei,Burkholderia mallei, andBurkholderia oklahomensis.J Antimicrob Chemother. 2020 May 1;75(5):1352-1353. 9. Zhang Chaodong#, Ju Yingjiao#, Tang Na#, Li Yun, Zhang Gang, Song Yuqin, Fang Hailing, Yang Liang*, Feng Jie*. Systematic analysis of supervised machine learning as an effective approach to predicate -lactam resistance phenotype inStreptococcus pneumoniae. Brief Bioinform. 2020 Jul 15;21(4):1347-1355. 10. Zhang Gang, Sun Kaiwen, Ai Guomin, Li Jianjuan, Tang Na, Wang Chao*, Feng Jie*. A novel family of intrinsic chloramphenicol acetyltransferase CATC inVibrio parahaemolyticus: naturally occurring variants reveal diverse resistance levels against chloramphenicol. Inter J Antimicrob Agents. 2019 Mar 13. pii: S0924-8579(19)30065-2. 11. Du Xiaochen#, Bayliss Sion C#,Feil Edward J, Liu Ying, Wang Chao, Zhang Gang, Zhou Dongsheng, Wei Dawei, Tang Na, Leclercq S bastien O, Feng Jie*. Real time monitoring of Aeromonas salmonicidaevolution in response to successive antibiotic therapies in a commercial fish farm. Environ Microbiol. 2019 Jan 13. doi: 10.1111/1462-2920.14531. 12. Tian Jingjing#, Zhang Gang#, Ju Y, Tang Na, Li Jianjuan, Jia Rufu, Feng Jie*. Five novel carbapenem-hydrolysing OXA-type -lactamase groups are intrinsic inAcinetobacterspp. J Antimicrob Chemother. 2018, 73(12):3279-3284. 13. Zhang Gang, Leclercq Sebastien Olivier, Tian Jingjing, Wang Chao, Yahara Koji, Ai Guomin, Liu Shuangjiang, Feng Jie*. A new subclass of intrinsic aminoglycoside nucleotidyltransferases, ANT(3")-II, is horizontally transferred among Acinetobacter spp. by homologous recombination. PLoS Genet. 2017 Feb 2; 13(2): e1006602. 14. Leclercq SO#, Wang C#, Sui Z, Wu H, Zhu B, Deng Y, Feng Jie*. A multiplayer game: species of Clostridium, Acinetobacter, and Pseudomonas are responsible for the persistence of antibiotic resistance genes in manure-treated soils. Environ Microbiol. 2016 Oct;18(10):3494-3508. 15. Zhang Gang, Tian Jingjing, Wang Chao, Chen Jifeng, Feng Jie*. Identification of novel cryptic aminoglycoside phosphotransferases in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 2016, 60(11):6983-6985. 16. Zhang Gang, Wang Chao, Sui Zhihai, Feng Jie*. Insights into the evolutionary trajectories of fluoroquinolone resistance in Streptococcus pneumoniae. J Antimicrob Chemother. 2015, 70(9): 2499-2506. 17. Wang Chao#, Sui Zhihai#, S bastien Olivier Leclercq, Zhang Gang, Zhao Meilin, Chen Weiqi, Feng Jie*. Functional characterization and phylogenetic analysis of acquired and intrinsic macrolide phosphotransferases in the Bacillus cereus group. Environ Microbiol. 2015, 17(5): 1560-1573. 18. Zhou Wenqing, Yao Kaihu, Zhang Gang, Yang Yonghong, Li Yun, Lv Yuan, Feng Jie*. Mechanism for transfer of transposon Tn2010 carrying macrolide resistance genes in Streptococcus pneumoniae and its effects on genome evolution. J Antimicrob Chemother. 2014, 69(6): 1470-1473. 19. Yang Jing#, Wang Chao#, Wu Jinyu, Liu Li, Zhang Gang, Feng Jie*. Characterization of a multiresistant mosaic plasmid from a fish farm sediment Exiguobacterium sp. isolate reveals aggregation of functional clinic-associated antibiotic resistance genes. Appl Environ Microb. 2014, 80(4): 1482-1488. 20. Zhang Gang#, TianWenyu#, Wang Chao, Feng Jie*. Identification of a novel resistance mutation in parE that confers high-level resistance to moxifloxacin in Streptococcus pneumoniae. J Antimicrob Chemother. 2012, 67(11): 2773-2774. |
研究中代表性图片
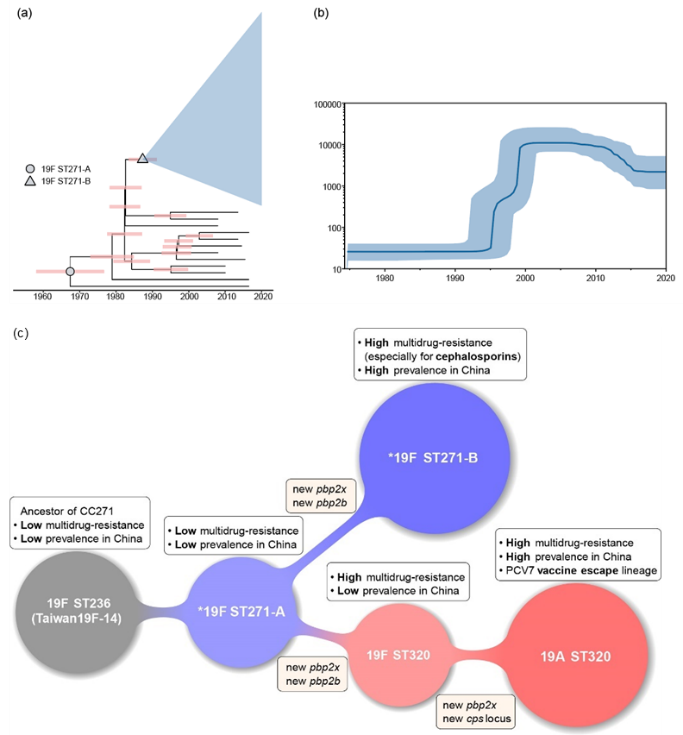
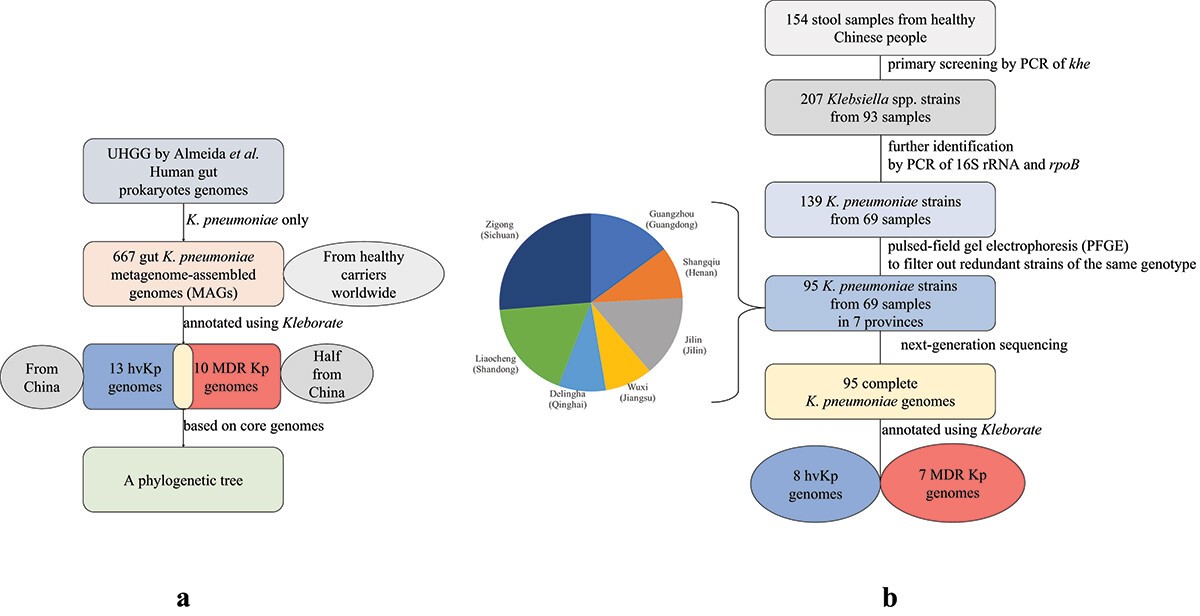
中国CC271克隆群的贝叶斯系统发育分析及主要群体演化历史示意图 (Genome Med. 2023 Jul 4;15(1):46) 健康人肠道可携带高毒力肺炎克雷伯菌 (Gut Microbes. 2022 Jan-Dec;14(1):2114739)
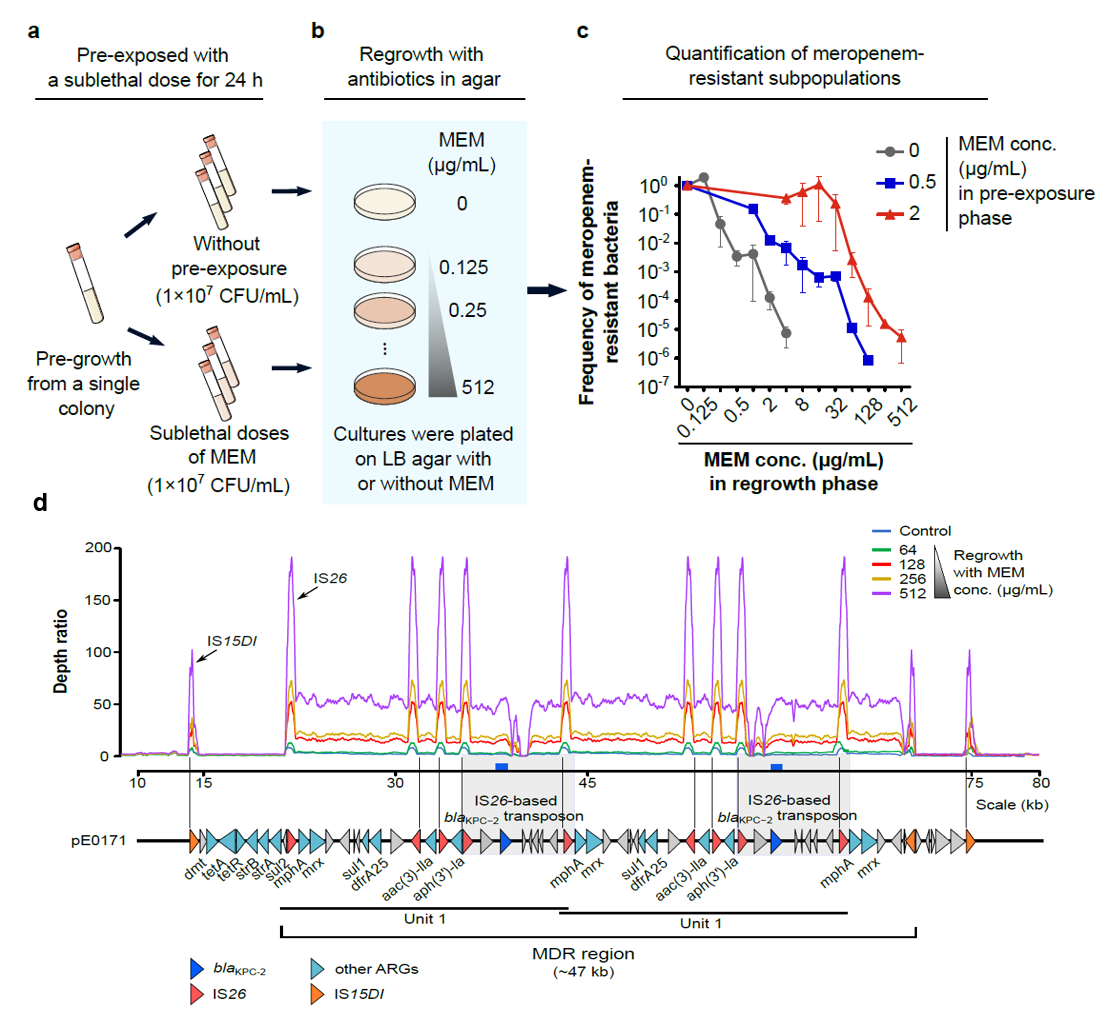
接触亚致死抗生素后菌株通过基因扩增迅速增加碳青霉烯耐药性 (mBio. 2022 Feb 8;13(1):e0334021.)
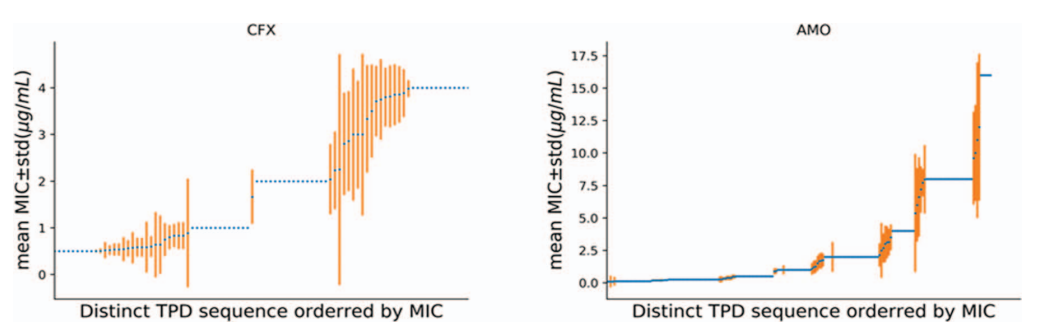
机器学习预测实验菌株的耐药水平 (Brief Bioinform. 2020 Jul 15;21(4):1347-1355.)
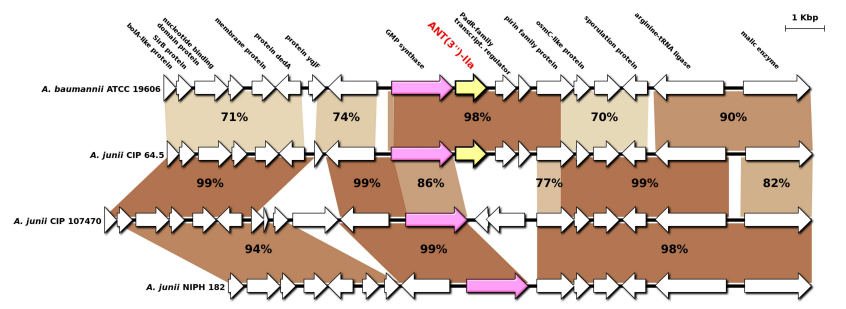
ant(3")-IIa基因的同源重组分析 (PLoS Genet. 2017 Feb 2; 13(2): e1006602) |